Rising rapidly, how did Apple develop cross-border AI models??
A protein folding model based on stream matching, SimpleFold, has been released and jokingly referred to as “iFold” by netizens.

SimpleFold does not have fancy exclusive module designs, but relies on a universal Transformer module, combined with a flow matching generation paradigm. The 3B parameter version rivals the performance of the top model in the field, Google AlphaFold 2.

Apple seems to be playing the game of simplifying complexity in this cross-border game.
##MacBook Pro runs effortlessly
Firstly, let’s talk about what protein folding is all about.
The core is to fold a “string” of amino acids into a specific 3D shape so that proteins can function effectively.
The protein folding model predicts the three-dimensional conformation of amino acids from their primary sequence.
The previous most powerful models, such as Google’s AlphaFold2, although achieved breakthroughs, used many complex proprietary designs.
For example, to analyze a large number of similar protein sequences, relying on multiple sequence comparisons (MSA) to construct evolutionary information, optimizing spatial constraints through triangular attention, and requiring the use of supercomputing power for inference, which is not affordable for ordinary laboratories.
But this’ iFold ‘solves this problem with a universal AI framework.
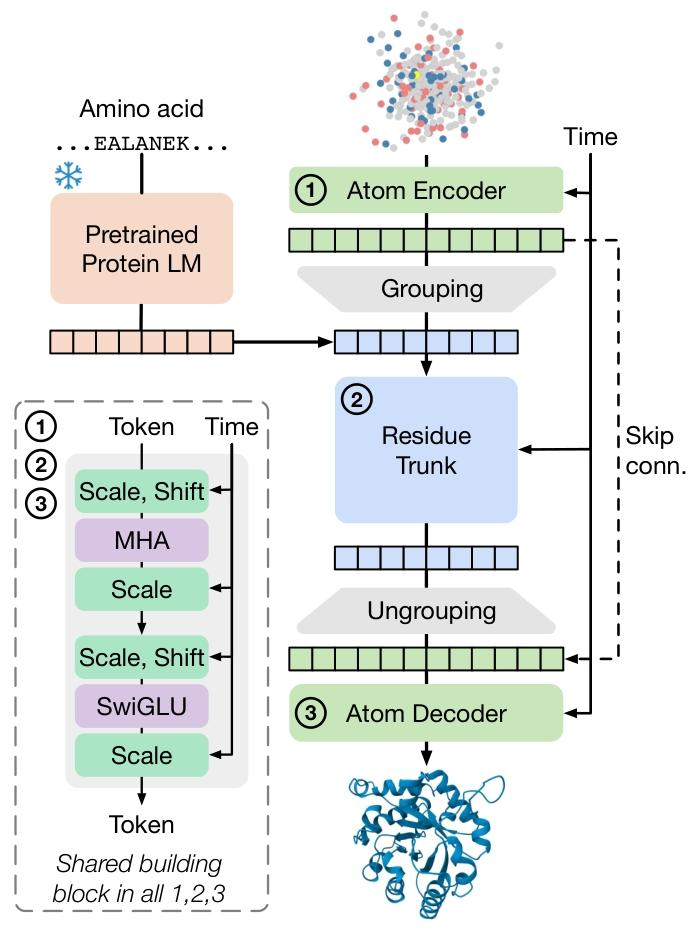
SimpleFold adopts a multi-layer Transformer encoder as the core backbone in its architecture, and only adapts protein sequence features through adaptive layer normalization, which is equivalent to using a “universal toolbox” to solve specialized domain problems.
The core innovation lies in the introduction of * * flow matching generation technology * *.
Unlike the gradual denoising of diffusion models, flow matching achieves one-step generation of atomic coordinates by learning a smooth mapping from random noise distribution to protein conformation distribution.
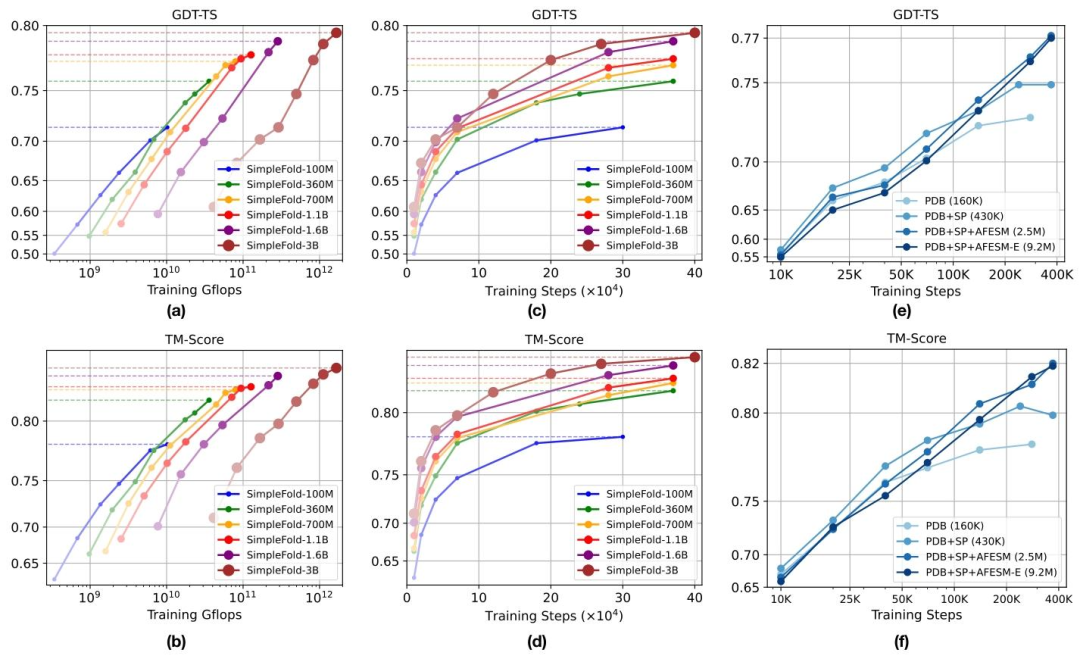
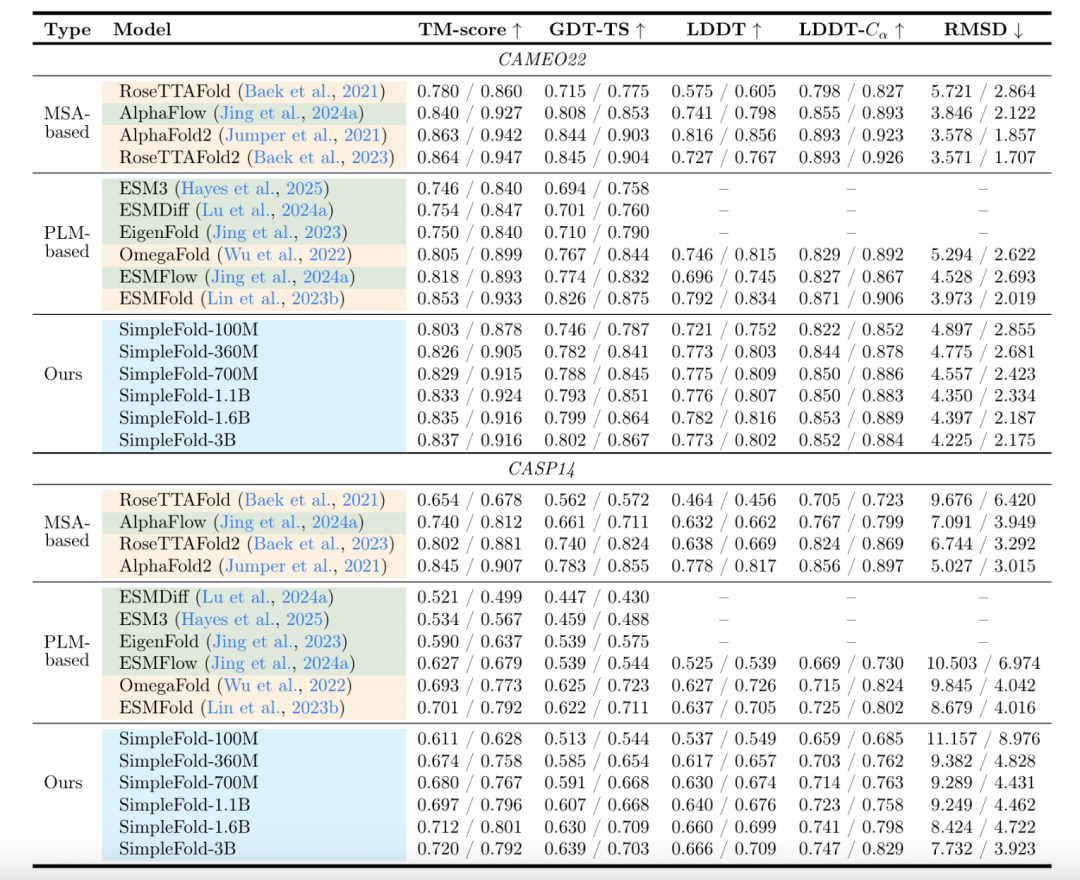
 During the training phase, the team constructed a mixed dataset containing 9 million pieces of data and trained a multi-scale model with 100M to 3B parameters. Among them, SimpleFold-3B achieved a performance of * * 95% * * of AlphaFold2 in the CAMEO22 benchmark test.
During the training phase, the team constructed a mixed dataset containing 9 million pieces of data and trained a multi-scale model with 100M to 3B parameters. Among them, SimpleFold-3B achieved a performance of * * 95% * * of AlphaFold2 in the CAMEO22 benchmark test.
On the CASP14 high difficulty test set, outperforms the ESMFold model for matching similar streams.
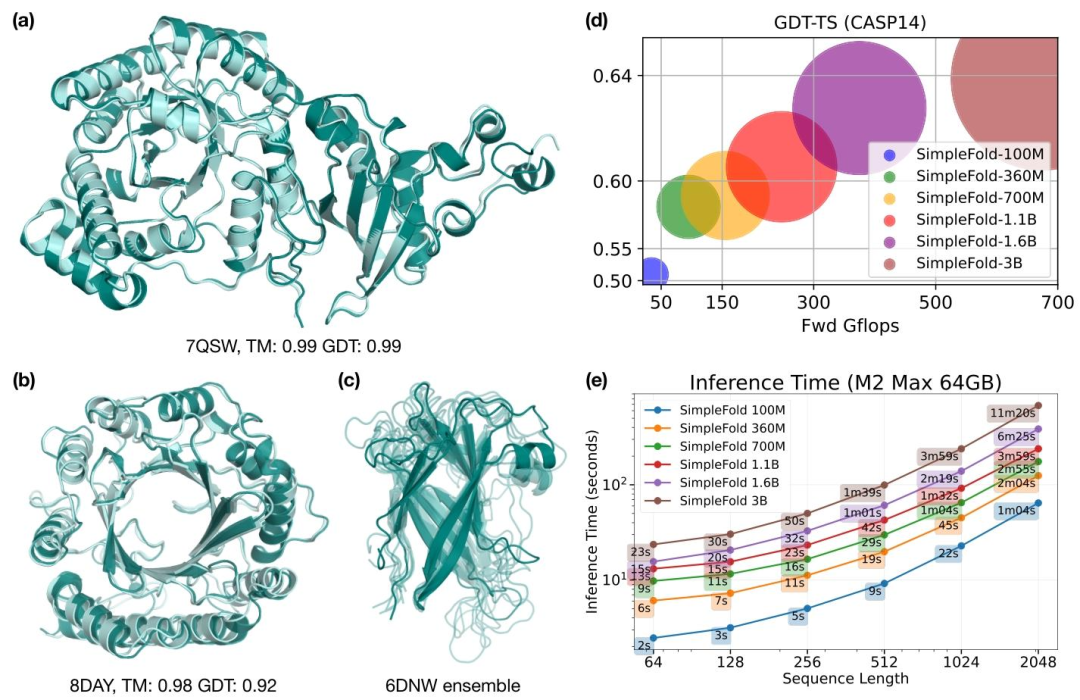
It is also worth mentioning the efficiency. On MacBook Pro equipped with M2 Max chip, the inference time for processing 512 residue sequences only takes two to three minutes, far exceeding the hourly time consumption of traditional models.
##Research Team
The first author of this study, * * Yuyang Wang * *, graduated from Tongji University with a bachelor’s degree and later went to Carnegie Mellon University in the United States for further studies. He obtained a master’s degree in mechanical engineering, a master’s degree in machine learning, and a doctoral degree in mechanical engineering, laying a solid foundation for his research in related fields through long-term studies.
He has internship experience in reinforcement learning development at Momenta and also worked as an AI/ML Resident at Apple, focusing on diffusion model research before becoming a machine learning researcher at Apple.

The corresponding author is Jiarui Lu, a Chinese machine learning engineer who graduated from Tsinghua University with a bachelor’s degree. During his studies, he also worked as a research assistant in Professor Zhu Jun’s laboratory.
Subsequently, Lu obtained a Master’s degree in Machine Learning from Carnegie Mellon University and joined Apple Inc. in 2020 after graduation.
I once led a benchmark for the ability to call large model tools - ToolSandbox, an open-source achievement of Apple.
For friends who are interested in this “iFold” and want to uncover technical details, please click on the link at the end of the article
*Paper address: https://arxiv.org/abs/2509.18480
Code address: https://github.com/apple/ml-simplefold *
*Reference link: [1] https://x.com/iScienceLuvr/status/1970787581248905454 *
